 Ground-breaking lab-on-a-chip research out of the van Nimwegen lab at the University of Basel, Switzerland and the Myers lab at the Max Planck Institute of Molecular Cell Biology and Genetics, Germany, has enabled both rapid environmental control of cell media, and the ability simultaneously monitor large numbers of bacteria as single cells. The work, recently published in Nature Communications and highlighted in Technology Networks, describes a PDMS microfluidic device with ~1900 channels measuring ~ 1 x 1 x 25 µm that house the single cells, and which are connected perpendicularly to larger supply channels containing the cell medium solution. The device also features continuously variable 2-input control of cell media via a split channel configuration, together with a mixing serpentine upstream of the smaller single cell channels. New image analysis software was also created to handle the enormous amount of image data generated by this system when in operation.
Ground-breaking lab-on-a-chip research out of the van Nimwegen lab at the University of Basel, Switzerland and the Myers lab at the Max Planck Institute of Molecular Cell Biology and Genetics, Germany, has enabled both rapid environmental control of cell media, and the ability simultaneously monitor large numbers of bacteria as single cells. The work, recently published in Nature Communications and highlighted in Technology Networks, describes a PDMS microfluidic device with ~1900 channels measuring ~ 1 x 1 x 25 µm that house the single cells, and which are connected perpendicularly to larger supply channels containing the cell medium solution. The device also features continuously variable 2-input control of cell media via a split channel configuration, together with a mixing serpentine upstream of the smaller single cell channels. New image analysis software was also created to handle the enormous amount of image data generated by this system when in operation.
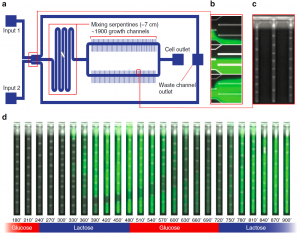
The device is intended to provide a real-time monitoring window into gene regulation in bacteria, a laudable advance. For example, bacterial response to different antibiotics can be measured, and insight into intra-cellular communication may be revealed. The device’s performance was nicely illustrated in a video (image above from Nature Communications article) showing an experiment where gene regulation via the lactose operon was observed while alternating between glucose and lactose nutrients in the supply channel; green fluorescent protein can be seen to appear within 1-2 h of introducing the lactose-rich medium to the cells.
