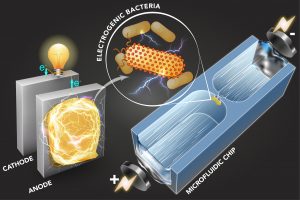
 New research out of Cullen Buie‘s lab at MIT’s LEMI lab shows effective sorting of different strains of bacteria using microfluidics-based dielectrophoretic (DEP) analysis. The research was published in Science Advances article (DOI: 10.1126/sciadv.aat5664) and also profiled by MIT News; a second MIT news article provides some background to microbial digestion and power generation.
New research out of Cullen Buie‘s lab at MIT’s LEMI lab shows effective sorting of different strains of bacteria using microfluidics-based dielectrophoretic (DEP) analysis. The research was published in Science Advances article (DOI: 10.1126/sciadv.aat5664) and also profiled by MIT News; a second MIT news article provides some background to microbial digestion and power generation.
The cell manipulation works by virtue of a DEP trap. A cell suspension flows through a small orifice (~ 50 µm wide) between two microfluidic chambers under an electric field. The electroosmotic and electrophoretic forces generated by the field determine each cell’s speed in the bulk solution. At the constriction, there is additionally a dielectrophoretic force that acts to trap cells of a certain polarisability, depending on the applied field strength. Since polarisability relates to extracellular electron transfer (EET), or a cell’s tendency to generate electricity during respiration, the trap can act as a filtre or screen for bacterial strains based on their tendency to generate electricity. In other words, different strains can be selectively trapped or screened based on the applied voltage.
The value of being able to discern between the different strains of bacterial in terms of their tendency to generate power while digesting the components in wastewater can hardly be understated. Apparently 3 of energy in the US is spent on wastewater treatment, while the wastewater itself contains 10 times the energy required for its own processing.
